|
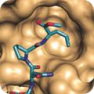 The interaction of proteins are critical to nearly all biological processes, including cellular signaling [1]. We offer special screening Peptidomimetic Libraries: β-Turn Peptidomimetic Library containing synthetic compounds which mimic beta-turns of proteins and a-Helix Peptidomimetic Library of compounds mimic alpha-helixes of proteins. These libraries are intended for research and drug discovery projects focused on protein-protein interactions. The interaction of proteins are critical to nearly all biological processes, including cellular signaling [1]. We offer special screening Peptidomimetic Libraries: β-Turn Peptidomimetic Library containing synthetic compounds which mimic beta-turns of proteins and a-Helix Peptidomimetic Library of compounds mimic alpha-helixes of proteins. These libraries are intended for research and drug discovery projects focused on protein-protein interactions.
 a-Helix Peptidomimetic Library with 1073 compounds was prepared Bayesian models using active template compounds from literature [2 - 8]. Two models were developed for each training sets. The first one was based on FCFP6 fingerprints, the second one – on ECFP6. Molecular descriptors, such as LogP, molecular weight, number of hydrogen donors and acceptors, number of rotatable bonds, number of rings and molecular polar surface area were involved in the construction of the models for more accuracy. a-Helix Peptidomimetic Library with 1073 compounds was prepared Bayesian models using active template compounds from literature [2 - 8]. Two models were developed for each training sets. The first one was based on FCFP6 fingerprints, the second one – on ECFP6. Molecular descriptors, such as LogP, molecular weight, number of hydrogen donors and acceptors, number of rotatable bonds, number of rings and molecular polar surface area were involved in the construction of the models for more accuracy.
 β-Turn Peptidomimetic Library (937 compounds in total) was prepared with two selection procedures and diversity clustering. The first selection was made using pharmacophore screening where the pharmacophore model was based on real β-turn structures [9]. The second selection was performed using similarity search to already known scaffolds of β-turn peptidomimetics such as pentameric [10] and hexameric [11] cycle scaffolds, bicyclic scaffolds [12] and others. β-Turn Peptidomimetic Library (937 compounds in total) was prepared with two selection procedures and diversity clustering. The first selection was made using pharmacophore screening where the pharmacophore model was based on real β-turn structures [9]. The second selection was performed using similarity search to already known scaffolds of β-turn peptidomimetics such as pentameric [10] and hexameric [11] cycle scaffolds, bicyclic scaffolds [12] and others.
Compounds with reactive groups and compounds containing any atom different to O, N, C, H, Br, I, Cl, F, P or S were removed from the libraries.
All compounds are in stock, cherry-picking is available. If your research is dedicated to PPI inhibitors, you also might be interested in our PPI Focused Libraries and Fragment Libraries.
The libraries (SDF, XLS, PDF format) are available on request. Feel free to contact us or use on-line form below to send an inquiry if you are interested to obtain these libraries or if you need more information.
-
Wesley E. , Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chem. Rev. – 1997 – Vol. 97, p. 1233−1250.
-
Maryanna E. Lanning and Steven Fletcher, Multi-Facial, Non-Peptidic α-Helix Mimetics. Biology 2015, 4, 540-555; doi:10.3390/biology4030540.
-
Christopher G Cummings and Andrew D Hamilton, Disrupting protein–protein interactions with non-peptidic, small molecule a-helix mimetics. Current Opinion in Chemical Biology 2010, 14:341–346
-
Jessica M. Davis, Lun K. Tsoub and Andrew D. Hamilton, Synthetic non-peptide mimetics of a-helices. Chem. Soc. Rev., 2007, 36, 326–334.
-
Madura K. P. Jayatunga, Sam Thompson, Andrew D. Hamilton, a-Helix mimetics: Outwards and upwards. Bioorganic & Medicinal Chemistry Letters 24 (2014) 717–724.
-
Maryanna Lanning & Steven Fletcher, Recapitulating the a-helix: nonpeptidic, low-molecular-weight ligands for the modulation of helix-mediated protein–protein interactions. Future Med. Chem. (2013) 5(18), 2157–2174.
-
Preethi Ravindranathan, Tae-Kyung Lee, Lin Yang, Margaret M. Centenera, Lisa Butler, Wayne D. Tilley, Jer-Tsong Hsieh, Jung-Mo Ahn & Ganesh V. Raj, Peptidomimetic targeting of critical androgen receptor–coregulator interactions in prostate cancer. Nature Communications (2013) 4,1923
-
Andrew J. Wilson, Helix mimetics: Recent developments. Progress in Biophysics and Molecular Biology (2015), doi:10.1016/j.pbiomolbio.2015.05.001.
-
Jari J. Koivisto, Esa T. T. Kumpulainen and Ari M. P. Koskine, Conformational ensembles of flexible b-turn mimetics in DMSO-d6. Org. Biomol. Chem. – 2010 – Vol. 8, p. 2103–2116.
-
Landon R. Whitby, Yoshio Ando, Vincent Setola, Peter K. Vogt, Bryan L. Roth, and Dale L. Boger, Design, Synthesis, and Validation of a β-Turn Mimetic Library Targeting Protein-Protein and Peptide-Receptor Interactions. J. Am. Chem. Soc. – 2011 – Vol. 133, p. 10184–10194.
-
Ralph F. Hirschmann et al., The β-D-Glucose Scaffold as a β-Turn Mimetic. Acc Chem Res. – 2009 – Vol. 42(10), p. 1511–1520.
-
Adam Golebiowsk et al., Solid-Supported Synthesis of Putative Peptide β -Turn Mimetics via Ugi Reaction for Diketopiperazine Formation. J. Comb. Chem. – 2002 – Vol. 4, p. 584–590.
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS

 a-Helix Peptidomimetic Library with 1073 compounds was prepared Bayesian models using active template compounds from literature [2 - 8]. Two models were developed for each training sets. The first one was based on FCFP6 fingerprints, the second one – on ECFP6. Molecular descriptors, such as LogP, molecular weight, number of hydrogen donors and acceptors, number of rotatable bonds, number of rings and molecular polar surface area were involved in the construction of the models for more accuracy.
a-Helix Peptidomimetic Library with 1073 compounds was prepared Bayesian models using active template compounds from literature [2 - 8]. Two models were developed for each training sets. The first one was based on FCFP6 fingerprints, the second one – on ECFP6. Molecular descriptors, such as LogP, molecular weight, number of hydrogen donors and acceptors, number of rotatable bonds, number of rings and molecular polar surface area were involved in the construction of the models for more accuracy. 
