|

The OTAVA GPCR Antagonist Library is a computationally designed collection created to explore GPCR-privileged chemical space with a focus on antagonist and inverse-agonist pharmacology.
The library integrates ligand-based machine learning, Bayesian modeling, and AI-driven scaffold optimization to enable efficient discovery of structurally diverse GPCR antagonists.
G protein-coupled receptors (GPCRs) represent the largest and most therapeutically validated class of drug targets in human biology. GPCR antagonists play a central role in suppressing pathological or excessive receptor signaling and are widely used across CNS, metabolic, cardiovascular, inflammatory, and respiratory indications.
Because GPCR antagonism depends on precise steric complementarity, controlled receptor occupancy, and balanced physicochemical properties, rational scaffold selection and diversity-driven library design are essential for successful hit discovery.
GPCR Target Class Significance
GPCR antagonists are essential for the treatment of diseases associated with receptor overactivation or dysregulated signaling, including:
-
Neurological and psychiatric disorders (e.g., schizophrenia, anxiety, addiction)
-
Cardiovascular diseases (hypertension, arrhythmias)
-
Metabolic disorders
-
Inflammatory and autoimmune conditions
-
Respiratory diseases (asthma, COPD)
-
Pain and sensory dysfunction
Due to the need for subtype selectivity, signaling pathway control, and minimization of off-target effects, discovery of novel, structurally diverse GPCR antagonists remains a high-priority challenge in drug development.
Library Design and Computational Workflow
1. Ligand-Based Machine Learning-Driven Antagonist Selection
The design of the GPCR Antagonist Library began with an integrated ligand-based machine learning workflow, combining multiple complementary modeling approaches to prioritize antagonist-like chemotypes.
A curated training collection of known GPCR antagonists and inactive compounds was used to construct predictive models, while a large screening collection derived from a proprietary ~300,000-compound stock served as the input chemical space.
The workflow incorporated k-NN classification, Bayesian probabilistic models, and iterative validation and ranking steps. Cross-validation procedures were applied to ensure robustness and generalizability of predictions.
As a result of this multi-model ML screening and probabilistic ranking, 4,231 GPCR antagonist-likeness candidates were selected as an initial enrichment layer for subsequent AI-driven optimization.
2. AI-Driven Optimization and Scaffold-Centric Refinement
The Bayesian- and ML-enriched antagonist candidates were further refined using AI-driven optimization, including XGBoost-based classification and similarity-guided prioritization.
Murcko scaffold extraction and clustering were applied to identify structurally representative cores and reduce redundancy while preserving chemical diversity.
As a result, an AI-optimized GPCR antagonist library of 2,573 compounds was generated, representing a scaffold-balanced, screening-ready chemical space optimized for antagonist pharmacology.
3. Structural Standardization and Feature Engineering
All compounds were standardized using a unified cheminformatics workflow, including:
-
Canonical SMILES generation and salt removal
-
Tautomer normalization and InChIKey-based deduplication
-
Murcko scaffold annotation and clustering
-
Physicochemical descriptor calculation (MolWt, cLogP, TPSA, HBA/HBD, RotB, QED)
-
PAINS and reactivity filtering
This ensured consistent, machine-learning-ready data representation and reliable downstream analysis.
4. Machine-Learning Scoring and Candidate Prioritization
The refined chemical space was evaluated using XGBoost-based machine learning models, integrating:
-
Structural similarity to known GPCR antagonist chemotypes
-
Scaffold-level diversity constraints
-
Favorable physicochemical ranges characteristic of orally active GPCR antagonists
This probabilistic ranking strategy enabled selection of high-quality candidates while maintaining broad scaffold coverage and avoiding over-concentration on narrow chemotypes.
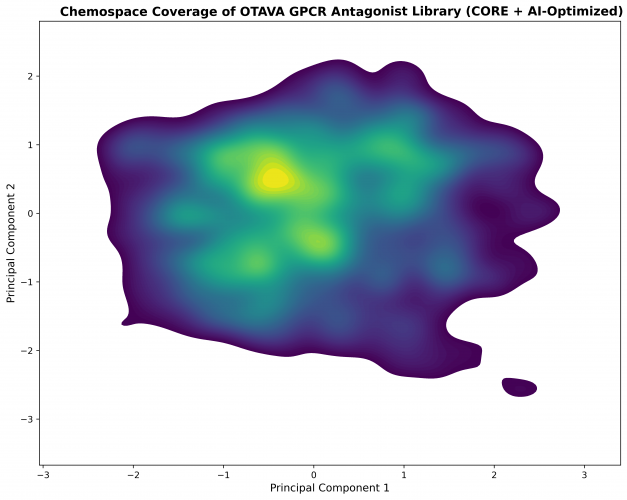
Scaffold Diversity and Chemospace Coverage
Principal component analysis (PCA) of the AI-optimized GPCR antagonist library demonstrates:
-
A dense central GPCR-relevant region enriched in heteroaromatic and fused ring systems
-
Multiple peripheral clusters capturing alternative binding modes, rigid and semi-rigid frameworks, and unique aromatic architectures
-
Smooth, continuous coverage of GPCR-relevant chemical space without sparsity gaps
This scaffold-balanced distribution enables complementary SAR exploration across multiple GPCR families and antagonist binding modes.
All the compounds are in stock, cherry-picking is available.
The libraries (DB, SD, XLS, PDF format) as well as the price-list are available on request. Feel free to contact us or use on-line form below to send an inquiry if you are interested to obtain this library or if you need more information.
Request Your Library Today! Fill out the form:
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS